Twist Article | Identifying Coronavirus Virulence Factors for RNA Vaccine Development with Twist Oligo Pools
The world continues to feel the impact of the global pandemic caused by SARS-CoV-2, the virus that results in COVID-19. It has undoubtedly changed life on earth; but as we continue to fight, new signs of progress give us hope. The research community has been particularly impacted and scientists have quickly mobilized to address and answer questions related to just how this virus infects, transmits, and mutates. One such group, profiled here, shows how Twist Oligo Pools can be used to drive exciting innovations, leading us closer to understanding how SARS-CoV-2 hijacks our immune system.
To cause infection, COVID-19 must enter a host cell and release its RNA genome. Virulence is established via replication of this genome, and production of new viral particles through the host cell’s expression of viral genes (Boopathi et al., 2020). Our cells have a number of innate mechanisms that restrict viral replication including RNA degrading proteins and genetic checkpoints alerting the immune system that “this cell has been infected.” These systems work in tandem with the adaptive immune system to halt virulence.
Yet, in the arms-race for survival, some viruses have evolved resistance to our innate immune systems. In some RNA viruses, the stability of their genome affords protection from RNA degrading enzymes. RNA stability is increased when it forms complex secondary structures through base-pairing with itself. These secondary structures can be unsusceptible to, or even actively block the host-cell’s antiviral ordinance allowing the virus to replicate unnoticed.
Therapeutics that target the stability of quickly-evolving viral RNA may allow the host immune system to effectively degrade and clear viral infection. In a recently published article in Biochemical and Biophysical Research Communications, Wakida et al. developed a method to expansively interrogate RNA sequences derived from RNA and DNA viruses which contribute to RNA stability in host cells. Senior author Professor Nobuyoshi Akimitsu describes this study as a step toward enhancing RNA medicine.
“mRNA is unstable and translation in the cell is often poor. By studying natural elements that enhance the stability and structure of mRNA, we hope to enhance the functionality of mRNA as a medicine,” Prof. Akimitsu explained.
The method developed in this study is termed “Fate-seq”. Fate-seq works on the principle that stable RNA sequences will be degraded in a mammalian cell much slower than unstable sequences. By attaching RNA sequences of interest to a signal molecule like green fluorescent protein and introducing them into cells, degradation can be directly measured. Cells exhibiting the strongest fluorescent signal over time will contain reporters attached to the most stable RNA structures. Sequencing cells with the strongest signal then allows precise mapping of highly stable RNA sequences.
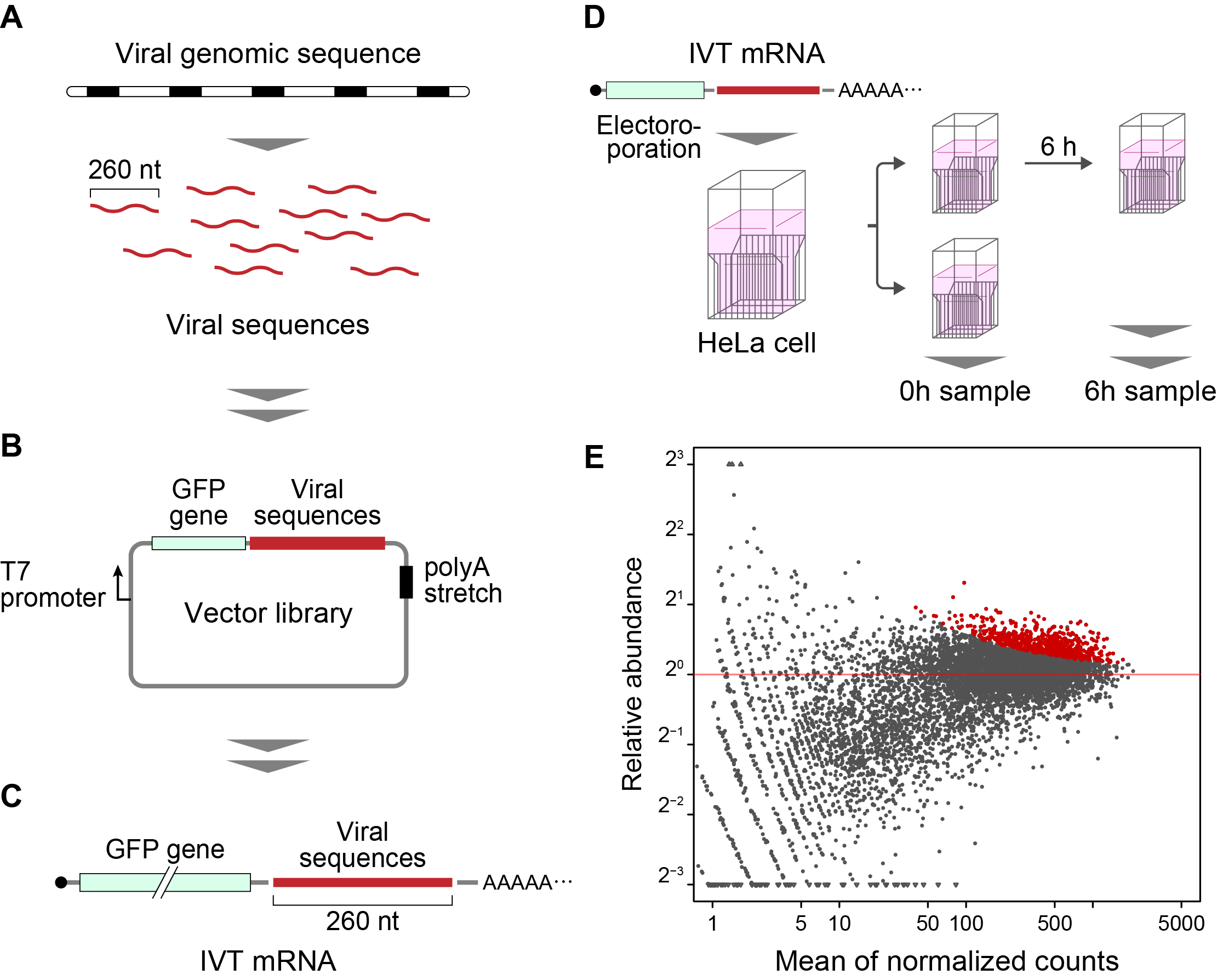
Wakida et al. used Twist Bioscience Oligo Pools to scan across the genome of 26 different viruses. Each viral genome was split into chunks of 260 nucleotides(nt), with 200nt overlap. Prof. Akimitsu described the importance of using high-quality oligos in the study.
“When doing a genome scan like Fate-seq, you need to be sure you are covering as much of the genome as possible, as evenly as possible. Because of the good uniformity and low error of Twist Oligo Pools, we can get even coverage across the viral genomes, and know we do not miss any potentially critical data.” Twist Oligo Pools can be synthesized up to 300 nucleotides, with an industry leading error rate of 1:2000.
In the study, Fate-seq uncovered two highly-conserved RNA stabilizing regions (COV001 and COV002) in severe acute respiratory syndrome-related coronavirus (SARS-CoV) and SARS-CoV-2. Results from Wakida et al. determined a change in secondary structure of COV001 in SARS-CoV-2 that was not inferred for SARS-CoV. Authors interpret that “this potential difference in the secondary structure may contribute toward the virulence of SARS-CoV-2. The identified sequences, which might affect viral RNA stability and RNA-binding proteins binding to these sequences, may offer possible targets for the treatment of viral infectious diseases including COVID-19.”
Development of COVID-19 vaccines are currently in high-demand, many consisting of messenger RNA to activate the host immune system (Callaway, 2020). New technologies like Fate-seq to identify stabilizing molecules will offer significant benefits to the RNA vaccine development field, allowing more stable and efficacious vaccines to be produced. Moreover, Prof. Akimitsu reiterated “RNA stability is important for the viral life cycle. In theory, by inhibiting stability, we can reduce viral proliferation and virulence.” Fate-seq is therefore an expeditious and promising approach to spotlight critical genomic regions for further characterization as targets for antiviral therapeutics.



