
Twist Bioscience NGS | Reagents & Kits | Universal Blockers
Twist Bioscience Next Generation Sequencing Reagents & Kits
Universal Blockers
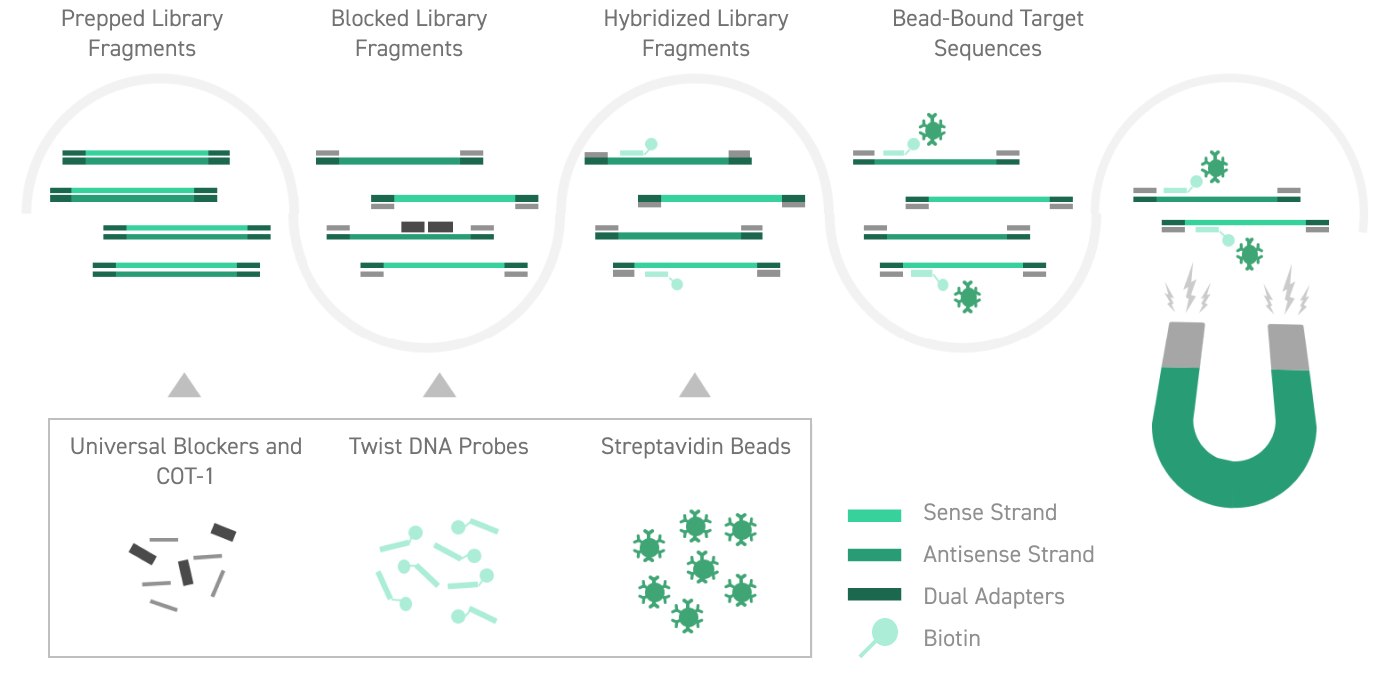
Optimizing the efficiency of next-generation sequencing (NGS) often involves targeted enrichment of genomic regions of interest prior to sequencing. In the Twist Bioscience target enrichment workflow, biotinylated synthetic DNA probes are designed to hybridize to exons or other custom genomic regions. Following hybridization with a genomic DNA sample, the probes are purified to produce a sample that is enriched for the targeted regions.
Twist Universal Blockers enhance the specificity of target capture by blocking non-specific hybridization between adapter sequences. Blocking non-specific interactions during hybridization simplifies experimental design and execution, reduces sequencing costs, and improves the results of targeted sequencing efforts.
KEY BENEFITS
- Prevent cross-hybridization between TruSeq-compatible adapters
- Enhance the performance of target enrichment workflows
- Blocks many different adapter designs and barcode lengths
- Effective across singleplex and multiplex target enrichment workflows
- Improve on-target capture regardless of panel size
- Ready-to-use formulation
- Reduce per-sample sequencing cost
- Simplify experimental design and execution
DATA
Block Off-Target Binding to Improve On-Target Capture
Twist Universal Blockers contain a proprietary mixture of universal adapter sequences1. These sequences bind genomic library fragments to block cross- hybridization between TruSeq-compatible adapters and improve on-target capture rates. Twist Universal Blockers can improve the performance of any target enrichment workflow that uses TruSeq-compatible adapters.
The robust design of Twist Universal Blockers maximizes flexibility for a more efficient target enrichment workflow. Compatible with all Twist Target Enrichment Panels, they improve capture of on-target reads independent of index sequence type, across singleplex and multiplex target enrichment workflows, and regardless of panel size. Twist Universal Blockers allow you the flexibility to choose the best adapter index design for your experiment without penalty to target enrichment performance.
Performance is independent of index design. On-target performance of Twist Universal Blockers across a variety of single and dual index TruSeq- compatible adapters. Individual libraries were generated from a single genomic source (NA12878; Coriell) and TruSeq-compatible adapters with index lengths ranging from 6 to 14 bp (unique molecular identifier [UMI] length not included). Hybrid capture was performed either in the absence or presence of Twist Universal Blockers using an exome target enrichment panel (33.1 Mb; Twist Bioscience) using 500 ng of gDNA per individual library following the manufacturer’s recommendations for 16-hour hybridization reactions. Cot DNA was present in all samples. Sequencing was performed with a NextSeq 500/550 High Output v2 kit to generate 2x76 paired end reads. Data was downsampled to 150x of target size and analyzed using Picard Metrics with a mapping quality of 20. Percentage of Bases On-Bait includes both On-Bait and Near-Bait Bases and is defined by the equation 1 - PCT_OFF_BAIT. Error bars denote one standard deviation or range of observations; N ≥ 2.
Performance is independent of panel size. On-target performance of Twist Universal Blockers across a variety of panel sizes. Individual libraries were generated from a single genomic source (NA12878; Coriell) and 8 bp dual-indexed TruSeq-compatible adapters. Hybrid capture was performed either in the absence or presence of Twist Universal Blockers with target enrichment panels of various sizes (0.04Mb to 33.1Mb; Twist Bioscience) and 500 ng of gDNA per individual library following the manufacturer’s recommendations for 16-hour hybridization reactions. Cot DNA was present in all samples. Sequencing was performed with a NextSeq 500/550 High Output v2 kit to generate 2x76 paired end reads. Data was downsampled to 150x of target size and analyzed using Picard Metrics with a mapping quality of 20. Percentage of Bases On-Bait includes both On-Bait and Near-Bait Bases and is defined by the equation 1 - PCT_OFF_BAIT. Error bars denote range of observations; N = 2.
SPECIFICATIONS
- Workflow Step: Target Enrichment
- Storage: Store at -5 to -30°C
SKUs
- 100856 - Twist Universal Blockers 2 Reactions
- 100578 - Twist Universal Blockers 12 Reactions
- 100767 - Twist Universal Blockers 96 Reactions
DOWNLOADS
Twist Universal Blockers Product Sheet - DOWNLOAD
* For research use only. Not for diagnostic procedures.



