
Twist Bioscience NGS | Library Preparation | Methylation Detection System
Twist Bioscience Next Generation Sequencing Library Preparation
NGS Methylation Detection System
The Twist NGS Methylation Detection System provides a robust, end-to-end sample preparation solution for identifying methylated regions in the human genome. The workflow employs a unique enzymatic process from New England Biolabs® that is much less damaging to DNA, alongside Twist‘s Custom Methylation Panel design.
Whether you are investigating cellular differentiation or screening liquid biopsies for cancer, the system offers the most efficient methylation detection available.
KEY BENEFITS
- State-of-the-art enzymatic conversion of unmethylated cytosines
- Uniform capture with high-performance custom Twist probes
- Improve on-target rates with optimized reagents
- Better results from higher library complexity and better GC balances
- Detects 15% more CpGs than bisulfite conversion
- Less sample damage enables challenging sample inputs
- Easily drop the workflow into existing Methyl-seq pipelines
- Adjust hybridization timing without sacrificing performance
- Improved on-target rates with Methylation Enhancer
- Hybridization in less than 4 hours
- Sophisticated design, accurate synthesis, and detailed QC maximize capture uniformity and reproducibility
- Capture sense, antisense, methylated and unmethylated DNA simultaneously
- Outstanding performance across panel sizes, target regions, and multiplexing requirements
- Easily add or enhance panel content
WEBINAR: Capabilities of the New Twist NGS Methylation Detection System - Watch Here
NEBNext® EM-seq™ Kit for Twist Targeted Methylation Sequencing
In partnership with New England Biolabs (NEB), Twist Bioscience offers a new methylation sequencing workflow that improves the quality of libraries and removes the need for damaging bisulfite treatment during prep.
The workflow features enzymatic conversion of unmethylated cytosines (see figure) to identify sites of methyl-cytosine (5mC) and hydroxymethyl-cytosine (5hmC).
Enzymatic conversion produces more intact libraries with better representation, and ultimately achieves more sensitive methylation detection. The library preparation system is suitable for whole genome sequencing and downstream enrichment with Twist Methylation Panels.
EM-seq conversion involves a series of enzymatic steps to convert unmethylated cytosines into uracils. The final result is the same as conventional bisulfite conversion, making EM-seq compatible with existing analysis pipelines that use Bismark and bwa-meth.
Twist Fast Hybridization System and Custom Methylation Panels
The Twist Fast Hybridization and Wash Kit provides optimal performance and maximum flexibility. Reagents and steps can be optimized to tune downstream sequencing metrics and to reduce hands-on time and pipette use.
Designed with a highly sophisticated algorithm, Twist Custom Methylation Panels capture targets with exceptional efficiency across a wide range of target sizes. Together, these components ensure best-in-class hybrid capture of custom regions of interest.
Twist Methylation Enhancer
Off-target capture frequently occurs in targeted methylation sequencing workflows because the conversion process reduces the sequence complexity of the sample during library preparation.
To maximize the capture of post-conversion samples, the Twist Methylation Detection System features a proprietary blocker designed specifically for methylation detection. Called Methylation Enhancer, this reagent reduces off-target capture by as much as half.
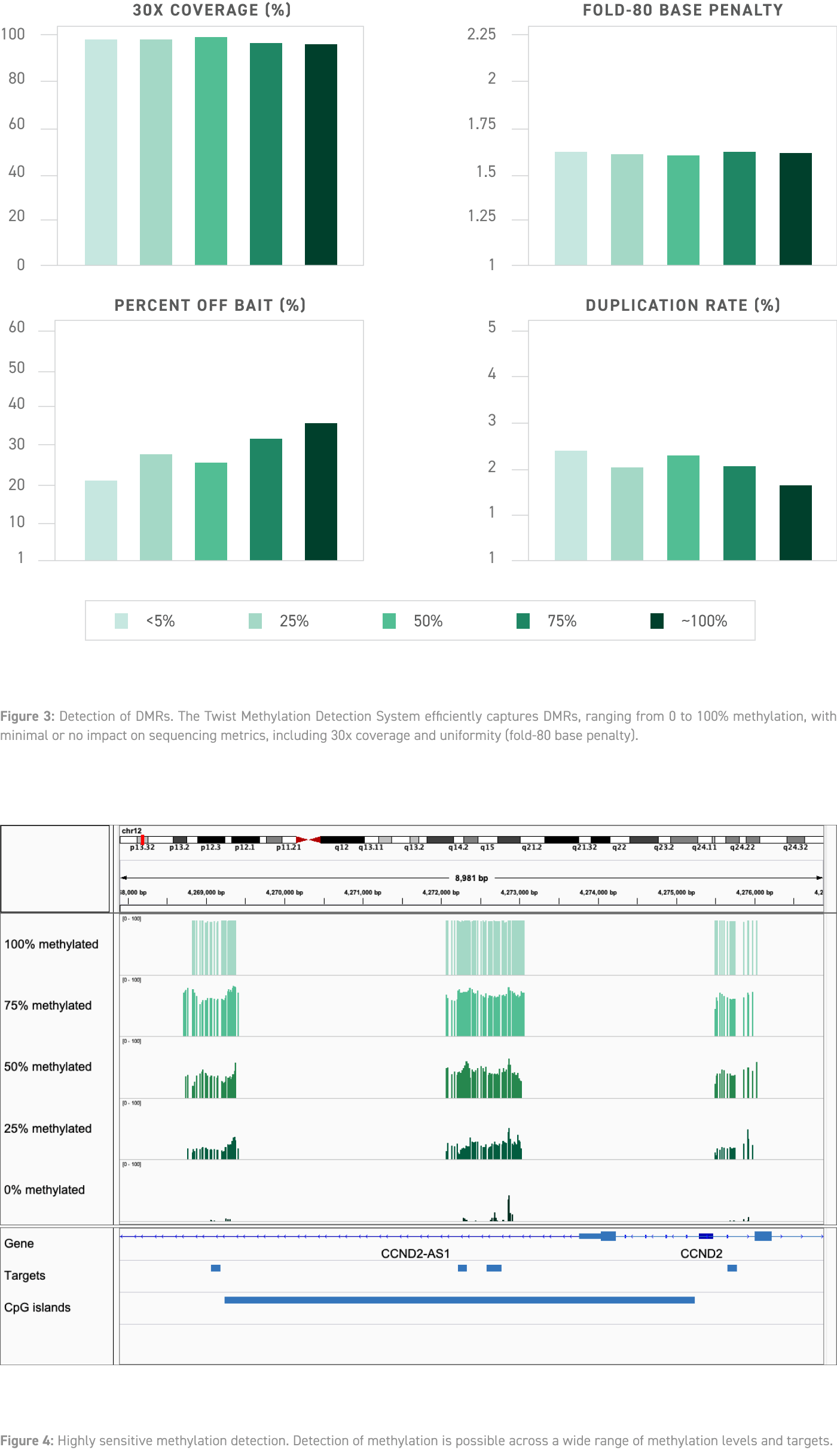
Highly Sensitive Detection of Differentially Methylated Regions
Methylation levels vary substantially across the human genome, and differentially methylated regions (DMRs) can be used to identify certain cancers.
To test the effects of methylation levels on the performance of the Twist Methylation Detection System, libraries of varying methylation levels (0-100% methylation) were generated by combining hypo- and hypermethylated genomic DNA in defined ratios. This analysis showed minimal effects of methylation level on final sequencing metrics.
The Twist Methylation Detection System also captures hypo- and hypermethylated regions with high sensitivity.
SKUs
- 101976 - NEBNext® EM-seq™ Kit for Twist Targeted Methylation Sequencing, 96 Samples
- 103557 - Twist Methylation Enhancer, 12 Reactions
- 100578 - Twist Universal Blocker, 12 Reactions
- 100983 - Twist Binding and Purification Beads Kit, 12 Reactions
- 101174 - Twist Fast Hybridization and Wash Kit, 12 Reactions
- 103496 - Twist Targeted Methylation Sequencing Workflow, 96x12 Reactions (contains all reagents listed above)
DOWNLOADS
- NGS Methylation Detection System - DOWNLOAD
- Highly Sensitive Methylation Detection Using Enzymatic Methyl-seq and Twist Target Enrichment - DOWNLOAD
- Minimizing Off-Target Capture in Targeted Methyl-Seq Through Panel Design & Enhancer Reagents - DOWNLOAD
- Optimizing the Performance of the Twist Targeted Methylation Sequencing Workflow - DOWNLOAD


